Note
Click here to download the full example code
Phosphene fading data from Perez Fornos et al. (2012)¶
This example shows how to use the Perez Fornos et al. (2012) dataset.
[PerezFornos2012] had nine Argus II users report perceived phosphene brightness via joystick position.
Important
You will need to install Pandas
(pip install pandas) for this dataset.
Loading the dataset¶
The dataset can be loaded as a Pandas DataFrame:
import numpy as np
import matplotlib.pyplot as plt
from pulse2percept.datasets import load_perezfornos2012
data = load_perezfornos2012()
print(data)
figure subject time_series
0 fig3_S1 S1 [nan, nan, nan, nan, 0.0, 10.289855072463762, ...
1 fig3_S2 S2 [nan, nan, nan, nan, 0.0, 9.456201550387616, 0...
2 fig3_S3 S3 [nan, nan, nan, nan, 0.0, 6.539635936582497, 9...
3 fig3_S4 S4 [nan, nan, nan, nan, 0.0, 2.759210526315784, 9...
4 fig3_S5 S5 [nan, nan, nan, nan, 0.0, 7.373643410852728, 9...
5 fig3_S6 S6 [nan, nan, nan, nan, 0.0, 9.053658536585358, 1...
6 fig3_S7 S7 [nan, nan, nan, nan, 0.0, 6.839285714285683, 1...
7 fig3_S8 S8 [nan, nan, nan, nan, 0.0, 9.966442953020133, 9...
8 fig3_S9 S9 [nan, nan, nan, nan, 0.0, 8.148230088495582, 4...
9 fig4_S1 S1 [nan, nan, nan, nan, 0.0, 9.583152173913025, 9...
10 fig4_S2 S2 [nan, nan, nan, nan, nan, 0.0885339538819553, ...
11 fig4_S3 S3 [nan, nan, nan, nan, 0.0, 9.355113636363637, 6...
12 fig4_S4 S4 [nan, nan, nan, nan, 0.0, 4.709610849056605, 9...
13 fig4_S5 S5 [nan, nan, nan, nan, 0.0, 5.867696323345818, 9...
14 fig4_S6 S6 [nan, nan, nan, nan, 0.0, 4.897842056932968, 9...
15 fig4_S7 S7 [nan, nan, nan, nan, 0.0, 10.423076923076923, ...
16 fig4_S8 S8 [nan, nan, nan, nan, 0.0, 9.825298540468818, 8...
17 fig4_s9 S9 [nan, nan, nan, nan, 0.0, 9.917355371900818, 7...
18 fig5_S1 S1 [nan, nan, nan, nan, 0.0, 6.46536796536796, 8....
19 fig5_S2 S2 [nan, nan, nan, nan, 0.0, 9.545454545454517, 0...
20 fig5_S3 S3 [nan, nan, nan, nan, 0.0, 9.252173913043483, 4...
21 fig5_S4 S4 [nan, nan, nan, nan, 0.0, 0.0, 0.5709745762711...
22 fig5_S5 S5 [nan, nan, nan, nan, 0.0, 6.098460336915207, 1...
23 fig5_S6 S6 [nan, nan, nan, nan, 0.0, 3.694806525659934, 9...
24 fig5_S7 S7 [nan, nan, nan, nan, 0.0, 7.210290722042516, 3...
25 fig5_S8 S8 [nan, nan, nan, nan, 0.0, 10.564581725133657, ...
26 fig5_S9 S9 [nan, nan, nan, nan, 0.0, 2.812601834084476, 7...
27 fig6_S1 S1 [nan, nan, nan, nan, 0.0, 9.566193845005886, 9...
28 fig6_S2 S2 [nan, nan, nan, nan, 0.0, 0.4496233521657302, ...
29 fig6_S3 S3 [nan, nan, nan, nan, nan, 7.515737410071948, 9...
30 fig6_S4 S4 [nan, nan, nan, -0.4626257135091045, -0.565371...
31 fig6_S5 S5 [nan, nan, nan, nan, nan, 7.978896103896103, 9...
32 fig6_S6 S6 [nan, nan, nan, nan, 0.0, 4.922984609602596, 6...
33 fig6_S7 S7 [nan, nan, nan, nan, 0.0, 5.908221351250469, 0...
34 fig6_S8 S8 [nan, nan, nan, nan, 0.0, 8.429239812152321, 9...
35 fig6_S9 S9 [nan, nan, nan, nan, 0.0, 6.600741633249903, 2...
36 fig7_S1 S1 [nan, nan, nan, nan, 0.0, 1.977431770468857, 3...
37 fig7_S2 S2 [nan, nan, nan, nan, 0.0, 2.0393053016453364, ...
38 fig7_S3 S3 [nan, nan, nan, nan, 0.0, 2.830743879472698, 6...
39 fig7_S4 S4 [nan, nan, nan, nan, 0.0, 1.4526049204052085, ...
40 fig7_S5 S5 [nan, nan, nan, nan, 0.0, 1.7023601398601456, ...
41 fig7_S6 S6 [nan, nan, nan, nan, 0.0, 1.719339622641511, 4...
42 fig7_S7 S7 [nan, nan, nan, nan, 0.0, 4.362728503723769, 8...
43 fig7_S8 S8 [nan, nan, nan, nan, 0.0, 3.976098776619857, 7...
44 fig7_S9 S9 [nan, nan, nan, nan, 0.0, 2.2775319567354897, ...
Inspecting the DataFrame tells us that there are 45 measurements (the rows) each with 3 different attributes (the columns).
These attributes include specifiers such as “subject”, “figure”, and “time_series”. We can print all column names using:
data.columns
Index(['figure', 'subject', 'time_series'], dtype='object')
Note
The meaning of all column names is explained in the docstring of
the load_fornos2012 function.
For example, “subject” corresponds to each subject used in the paper:
data.subject.unique()
array(['S1', 'S2', 'S3', ..., 'S7', 'S8', 'S9'], dtype=object)
To select all the rows where the subject frequency was ‘S2’, we can index into the DataFrame as follows:
print(data[data.subject == 'S2'])
figure subject time_series
1 fig3_S2 S2 [nan, nan, nan, nan, 0.0, 9.456201550387616, 0...
10 fig4_S2 S2 [nan, nan, nan, nan, nan, 0.0885339538819553, ...
19 fig5_S2 S2 [nan, nan, nan, nan, 0.0, 9.545454545454517, 0...
28 fig6_S2 S2 [nan, nan, nan, nan, 0.0, 0.4496233521657302, ...
37 fig7_S2 S2 [nan, nan, nan, nan, 0.0, 2.0393053016453364, ...
However, we can do the same operation when loading in the data as follows:
print(load_perezfornos2012(subjects='S2'))
figure subject time_series
0 fig3_S2 S2 [nan, nan, nan, nan, 0.0, 9.456201550387616, 0...
1 fig4_S2 S2 [nan, nan, nan, nan, nan, 0.0885339538819553, ...
2 fig5_S2 S2 [nan, nan, nan, nan, 0.0, 9.545454545454517, 0...
3 fig6_S2 S2 [nan, nan, nan, nan, 0.0, 0.4496233521657302, ...
4 fig7_S2 S2 [nan, nan, nan, nan, 0.0, 2.0393053016453364, ...
This leaves us with 4 rows, each pertaining to subject ‘S2’.
Note
Please see the documentation for
load_perezfornos2012 to see all
available parameters for data subset loading.
Plotting the data¶
One of the important points in the paper is how the perceptual response varies amongst subjects. Let us recreate Figure 3, for Subject 2, to show an example of this difference:
# Load in each figures data:
figure_3_s1_data = load_perezfornos2012(figures='fig3_S1')['time_series'][0]
figure_3_s2_data = load_perezfornos2012(figures='fig3_S2')['time_series'][0]
Stimuli were delivered over a 10 second window. To recreate this, we build
a vector of time points (time_steps) and set stimulation value to 10
within the first 10 seconds, and 0 otherwise:
time_steps = np.arange(start=-2.0, stop=75.5, step=0.5)
# Set up the pulse:
pulse_value = []
for pulse_step in time_steps:
if 10 >= pulse_step > 0:
pulse_value.append(10)
else:
pulse_value.append(0)
Now we can plot the stimulus alongside the subject’s response:
# Set up the subplot:
fig, (ax1, ax2) = plt.subplots(1, 2, sharex=True, sharey=True)
plt.xlim(-1, 30)
plt.ylim(-11, 11)
fig.suptitle('Average Joystick Response vs. Time')
# Plot subject 1
ax1.plot(time_steps, figure_3_s1_data, 'r', label='S1 Joystick Response')
ax1.step(time_steps, pulse_value, 'k--', label='Pulse')
ax1.spines['bottom'].set_position('center')
ax1.set_title('S1')
ax1.legend(loc='lower right')
# Plot subject 2
ax2.plot(time_steps, figure_3_s2_data, label='S2 Joystick Response')
ax2.step(time_steps, pulse_value, 'k--', label='Pulse')
ax2.spines['bottom'].set_position('center')
ax2.set_title('S2')
ax2.legend(loc='lower right')
fig.tight_layout()
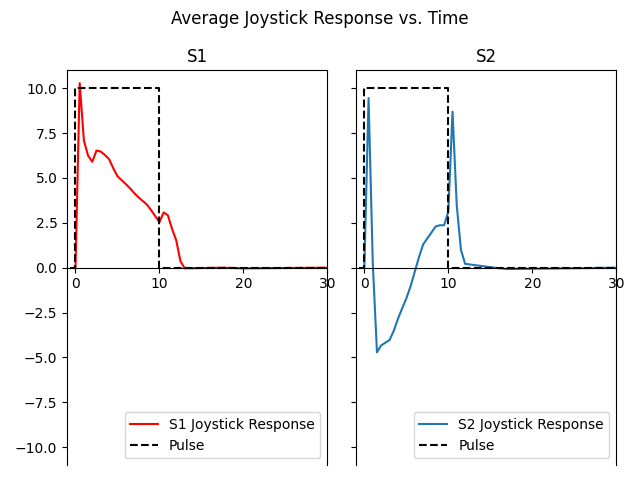
The above plot suggests that Subject 1 saw phosphenes gradually get dimmer over the stimulus duration. Once the stimulus was removed, phosphene brightness was quickly reduced to zero.
For Subject 2, the phosphene started bright (t=0) but rapidly went dark, even darker than the background (y-value < 0). Then the phosphene slowly got brighter again. Upon stimulus removal, the subject reported seeing another flash of light.
The reason for these differences is currently unknown, but might have to do with retinal adaptation.
Total running time of the script: ( 0 minutes 0.556 seconds)