Note
Click here to download the full example code
Data from Nanduri et al. (2012)¶
This example shows how to use the Nanduri et al. (2012) dataset.
[Nanduri2012] used a set of psychophysical detection tasks to determine size and brightness of phosphenes by modulating current amplitude and stimulating frequency in one Argus I user.
Important
You will need to install Pandas
(pip install pandas) for this dataset.
Loading the dataset¶
The dataset can be loaded as a Pandas DataFrame:
from pulse2percept.datasets import load_nanduri2012
data = load_nanduri2012()
print(data)
subject implant electrode ... interphase_dur pulse_type varied_param
0 S06 ArgusI A2 ... 0.45 cathodicfirst amp
1 S06 ArgusI A2 ... 0.45 cathodicfirst amp
2 S06 ArgusI A2 ... 0.45 cathodicfirst amp
3 S06 ArgusI A2 ... 0.45 cathodicfirst amp
4 S06 ArgusI A2 ... 0.45 cathodicfirst amp
.. ... ... ... ... ... ... ...
123 S06 ArgusI D4 ... 0.45 cathodicfirst amp
124 S06 ArgusI D4 ... 0.45 cathodicfirst amp
125 S06 ArgusI D4 ... 0.45 cathodicfirst amp
126 S06 ArgusI D4 ... 0.45 cathodicfirst amp
127 S06 ArgusI D4 ... 0.45 cathodicfirst amp
[128 rows x 17 columns]
Inspecting the DataFrame tells us that there are 128 measurements (the rows) each with 17 different attributes (the columns).
These attributes include specifiers such as “subject”, “electrode”, and “freq”. We can print all column names using:
data.columns
Index(['subject', 'implant', 'electrode', 'task', 'stim_class', 'stim_dur',
'freq', 'amp_factor', 'ref_stim_class', 'ref_amp_factor', 'ref_freq',
'brightness', 'size', 'pulse_dur', 'interphase_dur', 'pulse_type',
'varied_param'],
dtype='object')
Note
The meaning of all column names is explained in the docstring of
the load_nanduri2012 function.
For example, “freq” corresponds to the different stimulation frequency (hz) that were used in the paper:
data.freq.unique()
array([ 20., 15., 25., 40., 80., 120.])
To select all the rows where the stimulation frequency was 20hz, we can index into the DataFrame as follows:
print(data[data.freq == 20.0])
subject implant electrode ... interphase_dur pulse_type varied_param
0 S06 ArgusI A2 ... 0.45 cathodicfirst amp
1 S06 ArgusI A2 ... 0.45 cathodicfirst amp
2 S06 ArgusI A2 ... 0.45 cathodicfirst amp
3 S06 ArgusI A2 ... 0.45 cathodicfirst amp
4 S06 ArgusI A2 ... 0.45 cathodicfirst amp
.. ... ... ... ... ... ... ...
123 S06 ArgusI D4 ... 0.45 cathodicfirst amp
124 S06 ArgusI D4 ... 0.45 cathodicfirst amp
125 S06 ArgusI D4 ... 0.45 cathodicfirst amp
126 S06 ArgusI D4 ... 0.45 cathodicfirst amp
127 S06 ArgusI D4 ... 0.45 cathodicfirst amp
[88 rows x 17 columns]
This leaves us with 88 rows.
One of the important points of the paper is to investigate the relationship between phosphene brightness and size as either the stimulation amplitude factor or frequency varies. We can easily load in all data points where phosphene brightness was recorded when initially loading in the data set.
print(load_nanduri2012(task='rate'))
subject implant electrode ... interphase_dur pulse_type varied_param
0 S06 ArgusI A2 ... 0.45 cathodicfirst amp
1 S06 ArgusI A2 ... 0.45 cathodicfirst amp
2 S06 ArgusI A2 ... 0.45 cathodicfirst amp
3 S06 ArgusI A2 ... 0.45 cathodicfirst amp
4 S06 ArgusI A2 ... 0.45 cathodicfirst amp
.. ... ... ... ... ... ... ...
83 S06 ArgusI D4 ... 0.45 cathodicfirst freq
84 S06 ArgusI D4 ... 0.45 cathodicfirst freq
85 S06 ArgusI D4 ... 0.45 cathodicfirst freq
86 S06 ArgusI D4 ... 0.45 cathodicfirst freq
87 S06 ArgusI D4 ... 0.45 cathodicfirst freq
[88 rows x 17 columns]
Likewise, we can load in all data points where phosphene size was recorded when initially loading in the data set.
print(load_nanduri2012(task='size'))
subject implant electrode ... interphase_dur pulse_type varied_param
0 S06 ArgusI A2 ... 0.45 cathodicfirst amp
1 S06 ArgusI A2 ... 0.45 cathodicfirst amp
2 S06 ArgusI A2 ... 0.45 cathodicfirst amp
3 S06 ArgusI A2 ... 0.45 cathodicfirst amp
4 S06 ArgusI A2 ... 0.45 cathodicfirst amp
5 S06 ArgusI A4 ... 0.45 cathodicfirst amp
6 S06 ArgusI A4 ... 0.45 cathodicfirst amp
7 S06 ArgusI A4 ... 0.45 cathodicfirst amp
8 S06 ArgusI A4 ... 0.45 cathodicfirst amp
9 S06 ArgusI A4 ... 0.45 cathodicfirst amp
10 S06 ArgusI B1 ... 0.45 cathodicfirst amp
11 S06 ArgusI B1 ... 0.45 cathodicfirst amp
12 S06 ArgusI B1 ... 0.45 cathodicfirst amp
13 S06 ArgusI B1 ... 0.45 cathodicfirst amp
14 S06 ArgusI B1 ... 0.45 cathodicfirst amp
15 S06 ArgusI C1 ... 0.45 cathodicfirst amp
16 S06 ArgusI C1 ... 0.45 cathodicfirst amp
17 S06 ArgusI C1 ... 0.45 cathodicfirst amp
18 S06 ArgusI C1 ... 0.45 cathodicfirst amp
19 S06 ArgusI C1 ... 0.45 cathodicfirst amp
20 S06 ArgusI C4 ... 0.45 cathodicfirst amp
21 S06 ArgusI C4 ... 0.45 cathodicfirst amp
22 S06 ArgusI C4 ... 0.45 cathodicfirst amp
23 S06 ArgusI C4 ... 0.45 cathodicfirst amp
24 S06 ArgusI C4 ... 0.45 cathodicfirst amp
25 S06 ArgusI D2 ... 0.45 cathodicfirst amp
26 S06 ArgusI D2 ... 0.45 cathodicfirst amp
27 S06 ArgusI D2 ... 0.45 cathodicfirst amp
28 S06 ArgusI D2 ... 0.45 cathodicfirst amp
29 S06 ArgusI D2 ... 0.45 cathodicfirst amp
30 S06 ArgusI D3 ... 0.45 cathodicfirst amp
31 S06 ArgusI D3 ... 0.45 cathodicfirst amp
32 S06 ArgusI D3 ... 0.45 cathodicfirst amp
33 S06 ArgusI D3 ... 0.45 cathodicfirst amp
34 S06 ArgusI D3 ... 0.45 cathodicfirst amp
35 S06 ArgusI D4 ... 0.45 cathodicfirst amp
36 S06 ArgusI D4 ... 0.45 cathodicfirst amp
37 S06 ArgusI D4 ... 0.45 cathodicfirst amp
38 S06 ArgusI D4 ... 0.45 cathodicfirst amp
39 S06 ArgusI D4 ... 0.45 cathodicfirst amp
[40 rows x 17 columns]
Note
Please see the documentation for load_nanduri2012
to see all available parameters for data subset loading.
Plotting the data¶
To see the relationship between phosphene brightness as the amplitude factor varies,
we can recreate figure 4 a, from the paper.
Furthermore, the dataset available in load_nanduri2012
is used to create figures 4 and 5, a-d in the paper.
import matplotlib.pyplot as plt
import numpy as np
# load subset of the dataset concerning brightness data
brightness_data = load_nanduri2012(task='rate')
# get data where stimulation amplitude is varied
vary_amp = brightness_data[brightness_data.varied_param == 'amp']
# get the list of electrodes
electrodes = data['electrode'].unique()
# iterate over all electrodes
for electrode in electrodes:
# get relevant data for this specific electrode
electrode_data = vary_amp[vary_amp.electrode == electrode]
# normalize the amplitude
normalized_amp = electrode_data.amp_factor / electrode_data.ref_amp_factor
# set brightness rating
brightness_rating = electrode_data.brightness
# perform a first order linear best fit
linear_fit = np.poly1d(np.polyfit(normalized_amp, brightness_rating, 1))
# plot the linear best fit
plt.plot(normalized_amp, linear_fit(normalized_amp), label=electrode)
# display legend on plot
plt.legend()
# set plot axes
plt.xlim(0, 7)
plt.ylim(0, 60)
# set plot labels and title
plt.xlabel('Amplitude (uA) / Threshold (uA)')
plt.ylabel('Brightness Rating')
plt.title('Amplitude Modulation Brightness')
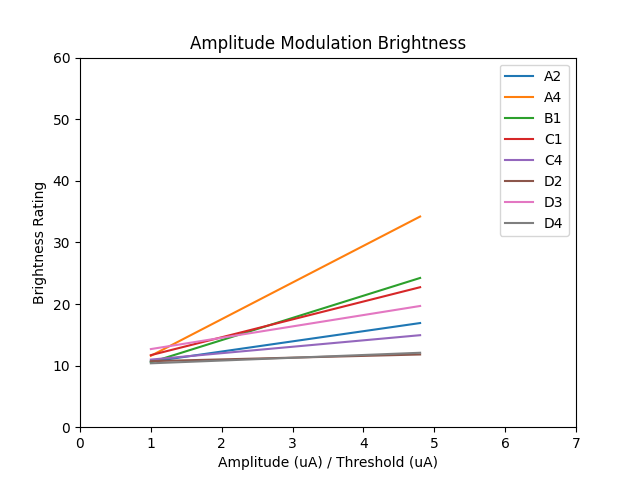
Text(0.5, 1.0, 'Amplitude Modulation Brightness')
Using Built-In Plotting Functionality¶
Arguably the most important column is “freq”. This is the current amplitude of the different stimuli (single pulse, pulse trains, etc.) used at threshold.
We might be interested in seeing how the phosphene brightness varies as a function of pulse frequency. We could either use Matplotlib to generate a scatter plot or use pulse2percept’s own visualization function:
from pulse2percept.viz import scatter_correlation
scatter_correlation(data.freq, data.brightness)
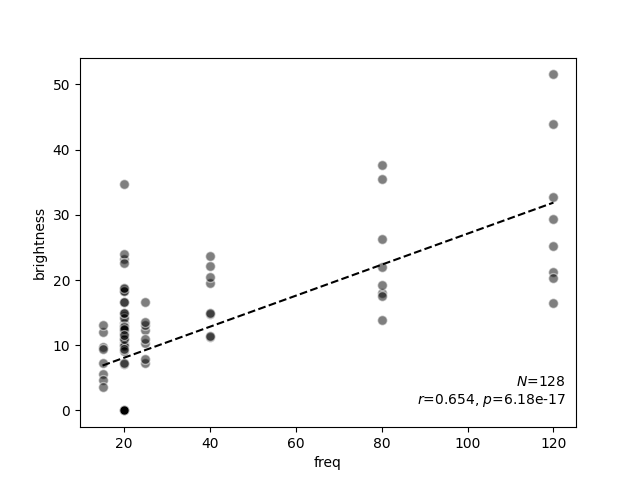
<Axes: xlabel='freq', ylabel='brightness'>
scatter_correlation above generates a scatter
plot of the phosphene brightness as a function of pulse frequency, and performs
linear regression to calculate a correlation $r$ and a $p$ value.
As expected from the literature, now it becomes evident that phosphene
brightness is positively correlated with pulse frequency
Total running time of the script: ( 0 minutes 0.420 seconds)